This page will host a growing repository of biosecurity risk mitigation tools developed as part of this programme. Most tools will become available in the second half of the research programme and any updates will be posted on the News page.
ExPAT (extended Pest Alert Tool)
The Pest Alert Tool is now significantly upgraded to ExPAT. Advances in high-throughput sequencing (HTS) technologies and their increasing affordability have fuelled worldwide application of environmental DNA (eDNA) metabarcoding data. The large volume of data produced by metabarcoding also raises the possibility of upscaling biosecurity surveillance capabilities, enabling incidental detection of species of concern through the broad assessment of entire biological communities. The tool has now two individual features:
1. Data Screening
2. Data Explorer
tombRaider - an algorithm to identify artefacts from metabarcoding data
Environmental DNA metabarcoding has revolutionized ecological surveys of natural systems. By amplifying and sequencing small gene fragments from environmental samples containing complex DNA mixtures, scientists are now capable of exploring biodiversity patterns across the tree of life in a time-efficient and cost-effective manner. However, the accuracy of species and haplotype identification can be compromised by sequence artefacts and pseudogenes. Despite various strategies developed over the years, effective removal of artefacts remains challenging and inconsistent data reporting standards hinder reproducibility in eDNA metabarcoding experiments. To address these issues, we introduce tombRaider, an open-source command line software program (https://github.com/gjeunen/tombRaider) and R package (https://github.com/gjeunen/tombRaider_R) to remove artefacts and pseudogenes from metabarcoding data post clustering and denoising. tombRaider features a modular algorithm capable of evaluating multiple criteria, including sequence similarity, co-occurrence patterns, taxonomic assignment, and the presence of stop codons. We validated tombRaider using various published data sets, including mock invertebrate communities, air eDNA from a zoo, and salmon haplotypes from aquatic eDNA. Our results demonstrate that tombRaider effectively removed a higher proportion of artefacts while retaining authentic sequences, thus enhancing the accuracy and reliability of eDNA-derived diversity metrics. This user-friendly software program not only improves data quality in eDNA metabarcoding studies, but also contributes to standardised reporting practices, an aspect currently lacking in this emerging research field.
Acknowledgement:
Funding:
Marsden Fast-Start MFP-UOO2116 (GJ)
New Zealand Ministry of Business, Innovation and Employment CAWX1904 (NG)
Level of Fouling (LOF)
The Level of Fouling (LOF) is a nominal rank scale with six categories and is widely used to rapidly characterize biofouling on submerged surfaces of ships and boats.
Auckland Council recently contracted Cawthron Institute’s Biosecurity Team to develop an updated guideline for application of the LOF scale. This document can be downloaded.
The LOF scale and its application will feature in some of the research associated with the Marine Biosecurity Toolbox programme.
CRABS software tool
With the establishment of next-generation sequencing technologies, an increasing amount of data can be generated in the pursuit to uncover diversity patterns and detect species of interest. However, the reliability and accuracy of classifying sequencing data depends on the algorithm used, as well as the quality and completeness of reference databases. While curated reference databases are available for commonly used genetic markers in microbial research, curated eukaryotic reference databases are scarce, as are the software tools for generating them. Here, we present CRABS (Creating Reference databases for Amplicon-Based Sequencing), a versatile tool for generating curated reference databases of user-specified genetic markers to aid taxonomy assignment from metagenomic and metabarcoding sequencing data. CRABS is available for download as a conda package and via GitHub.
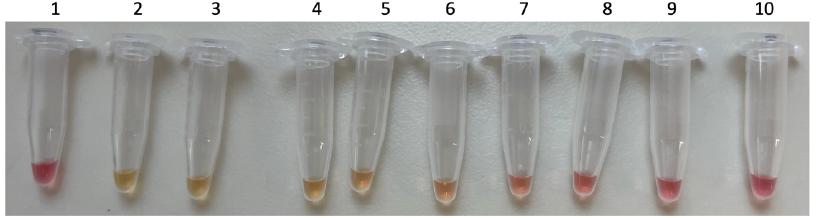
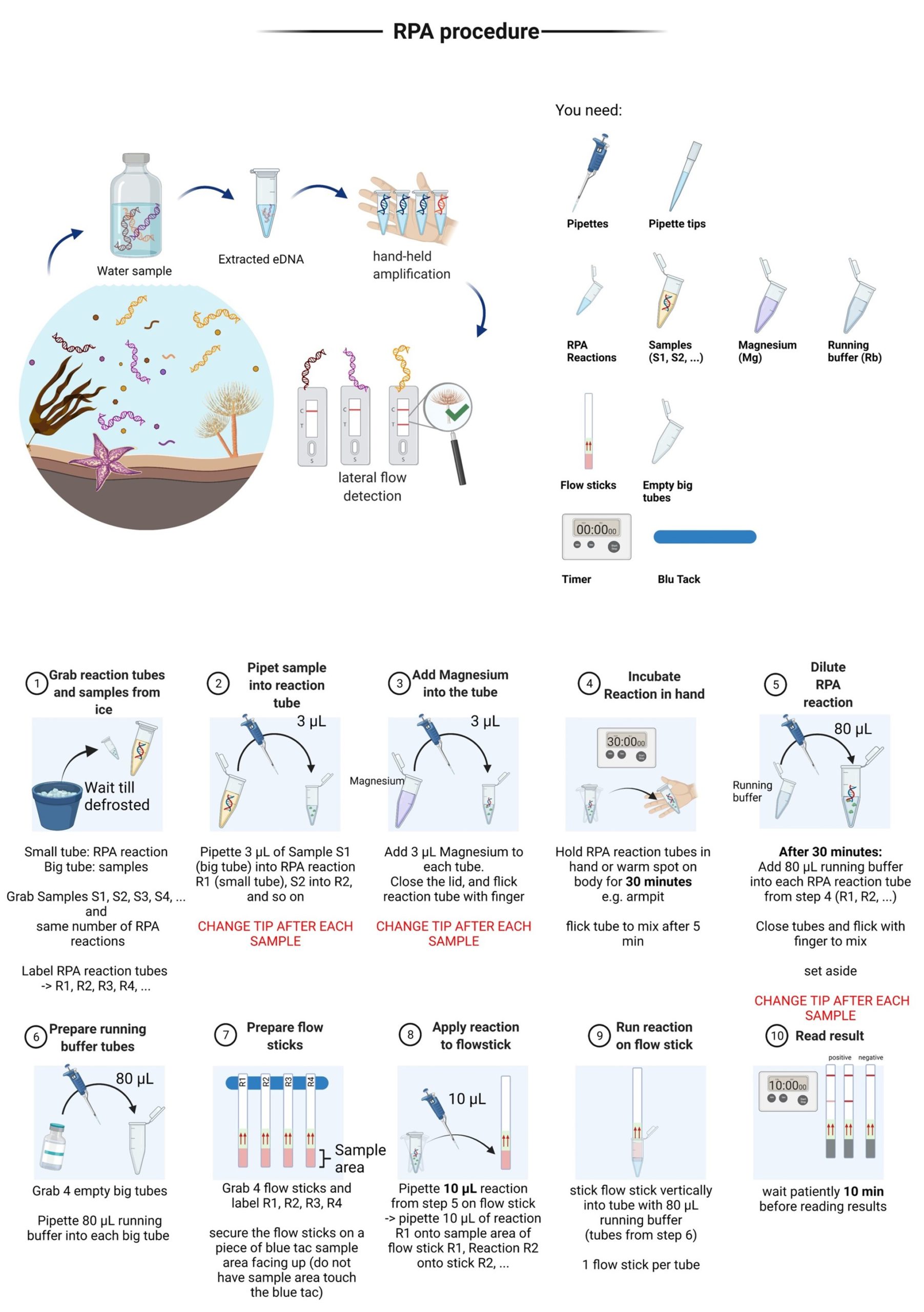
Simplified RPA-LF protocol for the Mediterranean fanworm detection
To address the current need in molecular surveillance techniques that are easy, fast, specific, and usable with minimal equipment and expertise, we have designed a recombinase polymerase amplification (RPA) assay for species-specific Sabella spallanzanii detection. Recombinase polymerase amplification is an isothermal amplification method, that amplifies target DNA within 20 minutes at a temperature range of 25°C to 45°C, which makes it possible to run assays at body temperature (~37°C), i.e., hand-held. With the introduction of labelled probes and primer, RPA can be used for fluorometry-based quantitative real time amplification. Detection is also possible with lateral flow (LF) strips, that provide easy visual read-outs. The combination of RPA and LF (RPA-LF) strip is especially valuable in resource-limited environments. RPA-LF was previously successfully implemented for multiple disease carrying organisms/viruses, including SARS-CoV-2 or for invasive species like the Northern Pacific Seastar, Asterias amurensis. A simplified RPA protocol was developed and tested with different end-user groups.
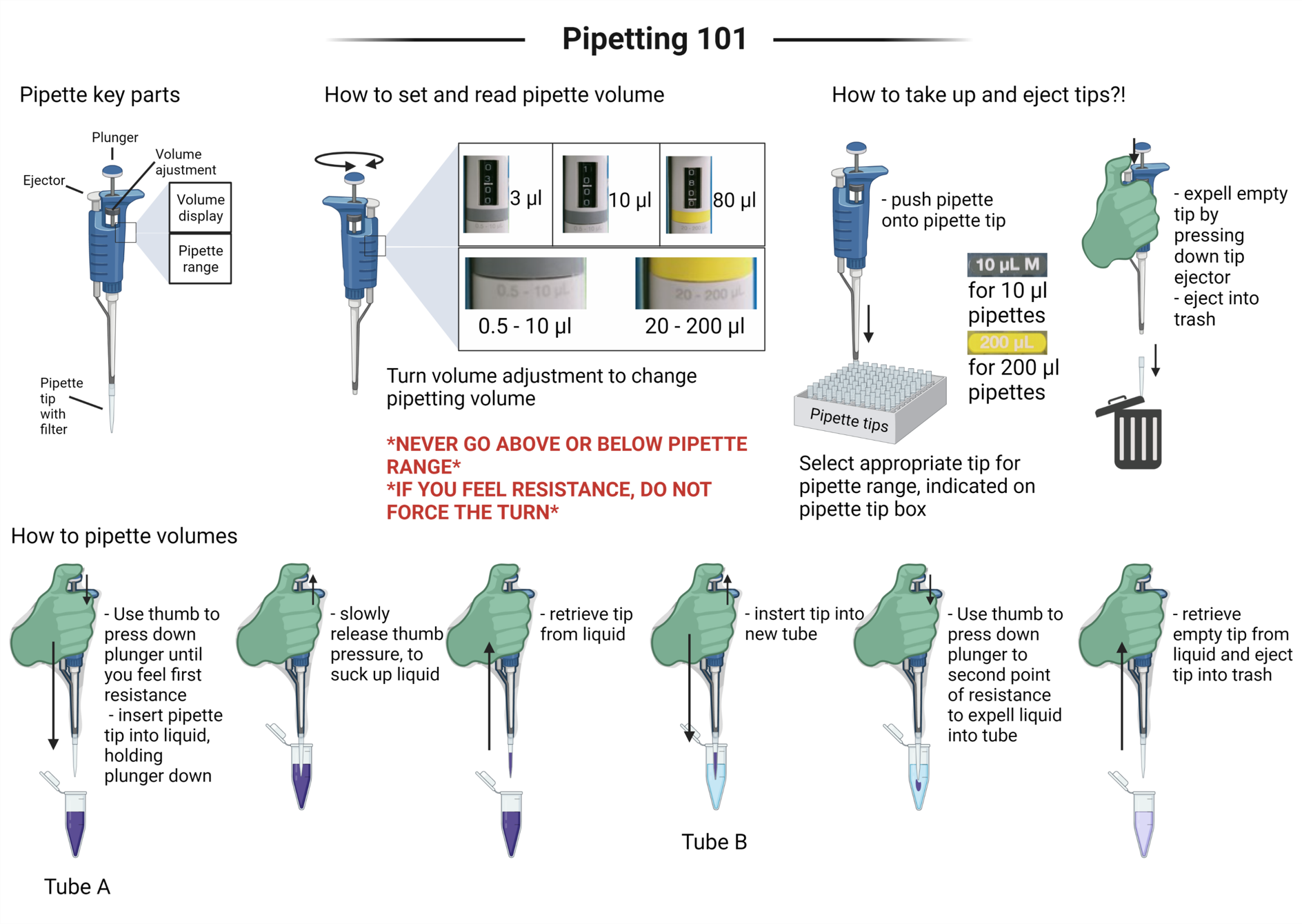
Pipetting 101 - a simplified guide for non-experienced users
At our outreach sessions and hands-on activities, we have noticed that micropipette usage is a hurdle for environmental DNA (eDNA) methods application by prospective biosecurity citizen scientists. To facilitate the induction process, we have created a visual pipetting guide, which can be used e.g. in a classroom by teachers when presenting molecular technologies, or added to a point-of-need molecular detection toolkit for biosecurity practitioners on the ground.
Environmental DNA best practice guidelines
The Environmental DNA Protocol Development Guide for Biomonitoring and Environmental DNA test validation guidelines were developed as a collaborative project, funded by the Australian Government Department of Agriculture, Fisheries and Forestry with support from the Marine Biosecurity Toolbox. More than 50 experts from across Australia and New Zealand were involved in the development of these guidelines and provided feedback during various stages of the process. These guidelines harmonise minimum quality standards to develop eDNA-based protocols and tests for biomonitoring applications. Both documents will be reviewed yearly to improve accuracy and clarity with current research and innovation.

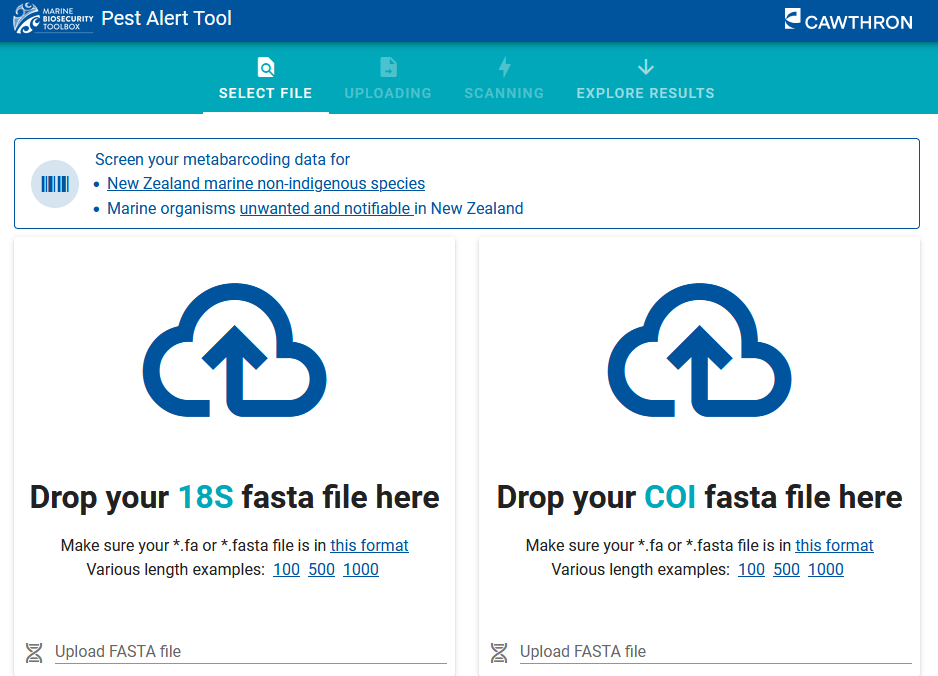
Pest Alert Tool
A new online tool called the Pest Alert Tool has been developed to screen high-throughput sequencing datasets for species of concern, such as non-indigenous and notifiable and unwanted organisms, in New Zealand’s marine environment, providing a valuable resource for biosecurity decision making. The tool can be adapted for other regions and applications and has the potential to be used for retrospective analysis of existing datasets, research purposes, and educational outreach activities.