This research theme focuses on non-indigenous species (NIS) that slip through the border and become introduced to New Zealand’s aquatic environments. Despite substantial biosecurity surveillance efforts, current approaches often fail to promptly detect risky species before they establish or spread too widely. This delay reduces opportunities for a successful response. This research theme will focus on developing and operationalising molecular surveillance technologies (i.e. sampling devices, designs and strategies) for biosecurity applications in natural, urban, and farmed marine environments.

Better sampling approaches:
when, where and how?
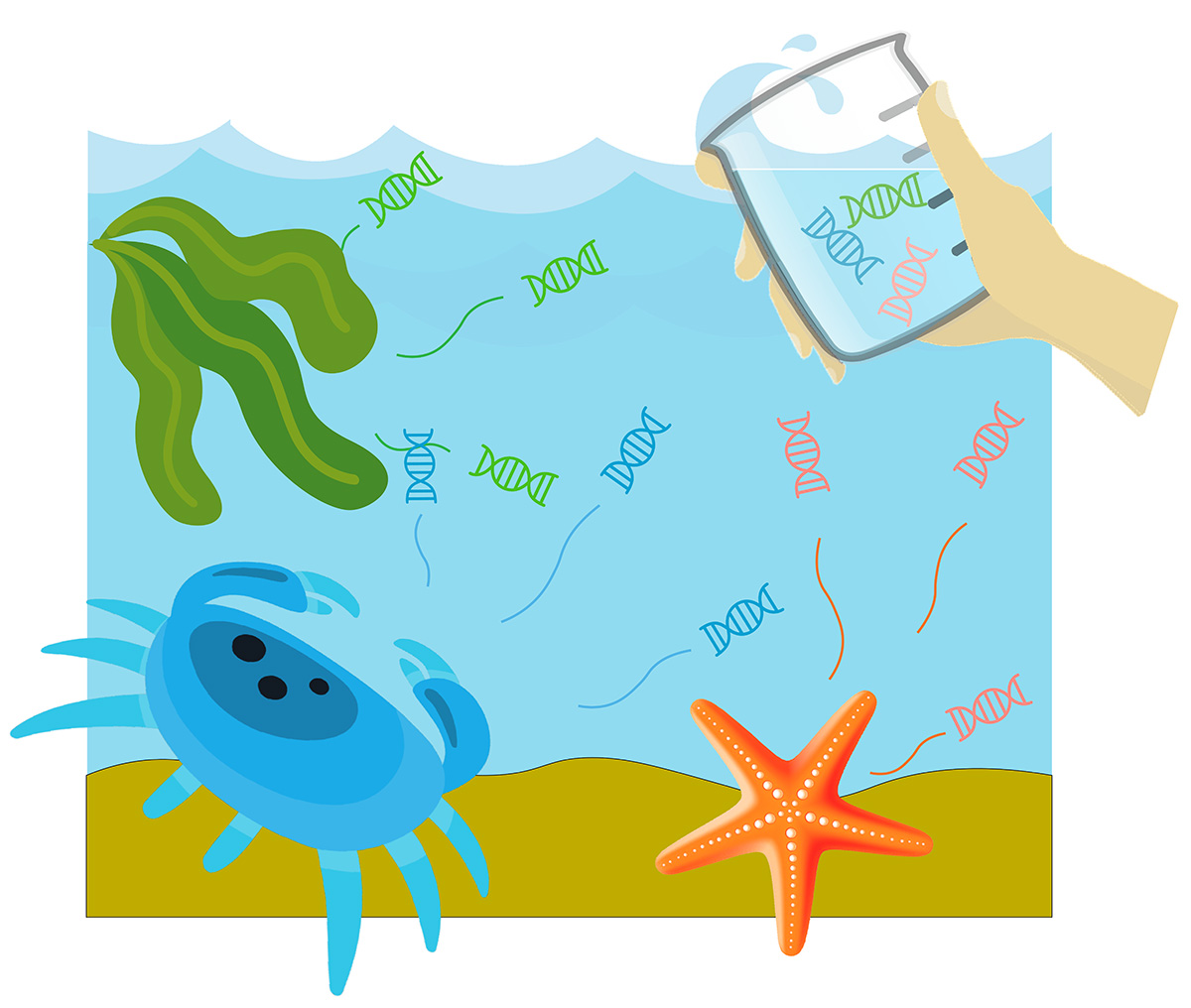
Marine organisms constantly release nucleic acids (DNA and RNA) into ambient environment in the form of free-floating molecules, particulate matter, expelled cells or living propagules (eggs and larvae). Environmental DNA and RNA (eDNA and eRNA) represent the organisms’ blueprint in an ecosystem and have proven useful for detection and identification of marine species, including those posing biosecurity risks. However, to harness the full potential of eDNA/eRNA-based methods for biosecurity, sampling approaches, designs and strategies must be optimised.
We will review existing methods for eDNA and eRNA isolation from aquatic environments and run a range of controlled experiments and field validations to assess their efficiency for biosecurity applications (both for general community screening and detection of target pests). We will employ modelling approaches to better understand dispersal of eDNA and eRNA in marine waters, enhance their detection probability by adjusting surveillance design (i.e. sampling times, locations, frequency) and predict where the source population is (i.e. where control or management efforts may need to be directed). The models will be parameterised with state-of-the-art empirical data on the fate of eDNA and eRNA fate in marine systems. Comprehensive cost-benefit analysis and proof testing of the surveillance designs via partnering councils’ operational biosecurity programmes and building upon the Mātauranga Māori (indigenous knowledge) of kaitiaki (guardians) in the regional environmental context, will guide the integration of these tools into routine biosecurity surveillance in New Zealand.
Updated toolkit for molecular biosecurity surveillance
With the rapid development of molecular technologies, new opportunities emerge for eDNA and eRNA biosecurity applications. We will explore the usability of alternative methodologies (including those from medicine or chemistry) for nucleic acid adsorption from water on functionalised substrates, cost-efficient preservation and isolation of eDNA and eRNA and field-based biodiversity screening tools. The most promising methods will be tested in mesocosm experiments and in situ, to assess their performance, sensitivity and specificity, and to identify potential sources of error and uncertainty that might result in false positives or non-detection of target species.
We will also develop new molecular assays for detecting marine pests of high priority to end-users, focusing on technologies easily adaptable for in situ applications (e.g. in field-deployable testing kits, portable or autonomously operating devices). In collaboration with international partners, we will be working on enhancing sequence reference databases for notorious marine pests, by producing and assembling high-quality data from newly sequenced metagenomes of selected pests. These data are imperative for the development and regional validation of species-specific assays, but also will help improve accuracy in identifying risky taxa from molecular biodiversity datasets. It will also help in unraveling the genetic variability of both native and non-indigenous marine organisms. Ultimately, this effort will feed into international genetic data repositories and automated molecular data screening (alert tools) for high-risk taxa in New Zealand.
Empowering citizen-based molecular biosecurity monitoring in New Zealand
We envisage that the molecular surveillance kits developed and operationalised in this programme will directly feed into citizen-science programmes and school curricula. We will work closely with two Nelson Colleges, social scientists and Māori partners to develop a framework for student engagement by introducing them to marine ecology, molecular biology, biosecurity and kaitiakitanga (guardianship for the nature) concepts. We will harness the creativity of young people to support the development of new molecular sampling tools, seek their feedback on prototypes developed during the programme and offer hands-on experience trialing these tools in the field as part of curricular science teaching and field trips. Bringing cutting-edge biosecurity science into NZ classrooms is key to developing the next generation of biosecurity scientists and end-users, in line with the vision of Biosecurity 2025 that focuses on ‘transforming the way we do biosecurity’ and engaging ‘a biosecurity team of 4.7 million‘. After our framework is tested and proven functional with a small focus group of students and teachers, wider dissemination will occur through educational sessions at other schools, iwi/hapū/whanau and kaitiaki, Science Centers and youth science conferences.
The research activities of the DETECT theme will be carried out in close collaboration with Patuharakeke Te Iwi Trust, Nelson College, Nelson College for Girls and the programme’s end-user partners (Northland, Auckland and Marlborough councils, Ministry for Primary Industries). Field-based trials and surveys will take place around Otago, Northland, Waitemata Harbour and the Top of the South region.