Last year over 700 boat owners who have non-trailered boats moored in marinas around Aotearoa…
Drop it all: extraction-free detection of targeted marine species through optimized direct droplet digital PCR
Recent research led by Ph.D. student Michelle Scriver from the DETECT team of the Marine Biosecurity Toolbox programme has been published in PeerJ. This publication showcases innovative collaborations and highlights the pioneering work conducted by scientists within the toolbox and partnering organizations.
In partnership with Nucleic Sensing Systems, our research team demonstrated the ability to detect species-specific marine environmental DNA (eDNA) directly from water samples, eliminating the need for extraction!
This innovative approach was showcased in a proof-of-concept aquarium study conducted at the Cawthron Institute, where we assessed the detectability of eDNA from marine invasive species using an optimized species-specific direct droplet digital PCR assay. Our results highlight factors influencing marine eDNA detectability with direct-ddPCR, paving the way for future advancements with this technology. This study lays the foundation for automated continuous monitoring and rapid management responses in marine environments. We are looking forward to future work and continued innovative technology to detect targeted species, such as rare or invasive species, further protecting and conserving our beautiful marine space.
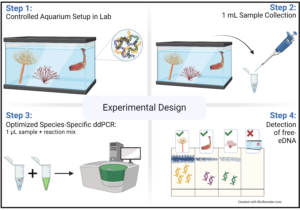
The experiment design: The schematic shows the four-step sampling procedure for detecting targeted environmental DNA (free-eDNA) with direct droplet digital polymerase chain reaction (direct-ddPCR). This figure was made in © BioRender: biorender.com.
The study was published March 2024 in Science of the Total Environment (STOTEN), read the whole publication here!




Comments (0)