Last year over 700 boat owners who have non-trailered boats moored in marinas around Aotearoa…
A rapid molecular assay for detecting the Mediterranean fanworm developed by DETECT team and trialed by non-scientist users
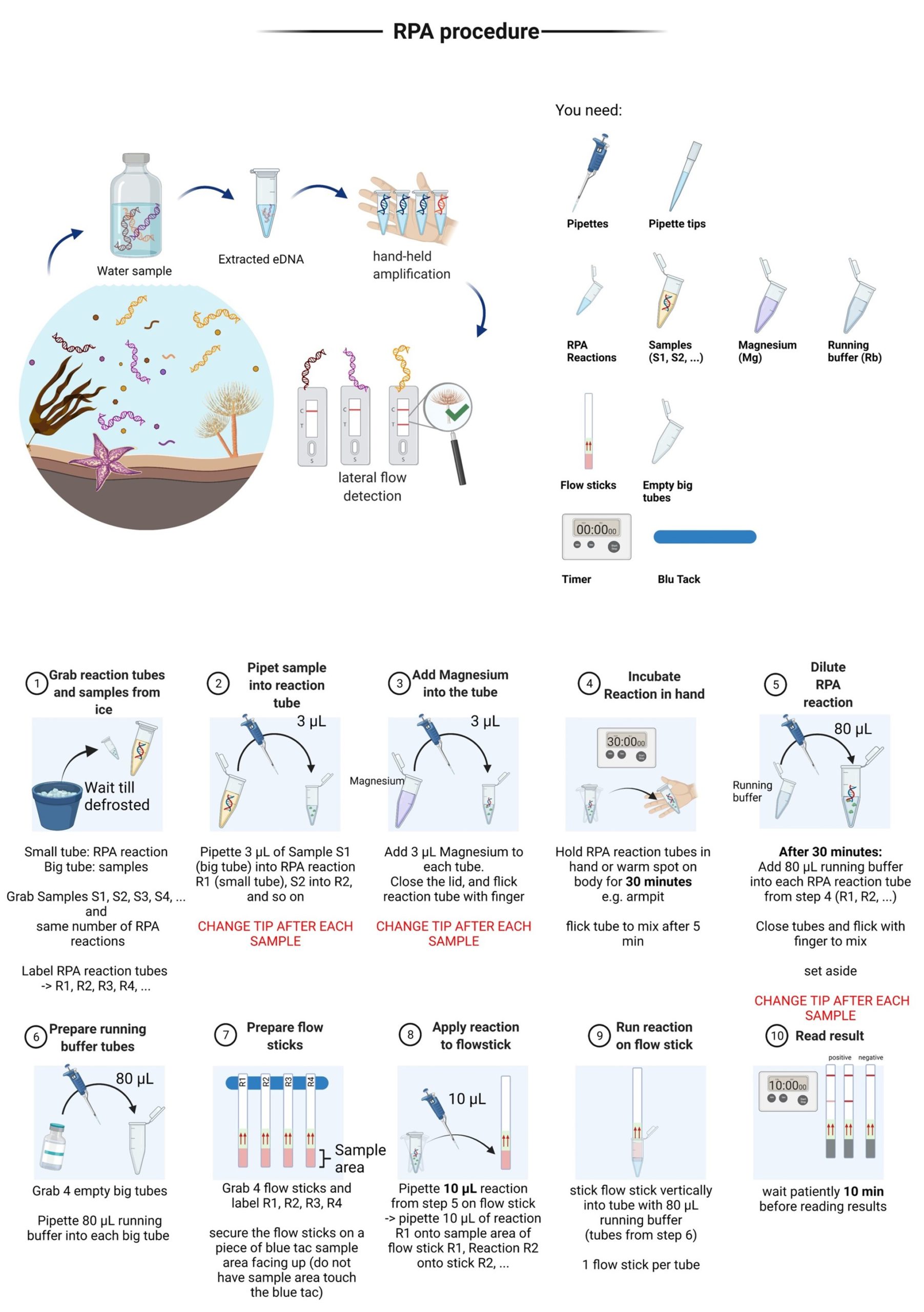
Sensitive molecular detection techniques gain great interest and are being increasingly tested for detecting the notorious fanworm in high-risk marine sites (i.e., ports and marinas) around New Zealand. However, conventional molecular detection assays require specific laboratory resources and technical expertise. This restricts the wider applicability of this approach by biosecurity practitioners or communities willing to be engaged in biosecurity surveillance. To provide end-users with a fast, easy and highly specific way to detect Sabella spallanzanii directly at the site of interest, a species-specific recombinase polymerase amplification (RPA) assay was designed to be read-out with lateral flow strips (RPA-LF). The RPA generates amplification within 20 minutes at 37-39°C, with a detection limit of 10 pg of the target DNA, matching the detection limit of digital droplet PCR (ddPCR) when performed on eDNA samples.
A simplified visual protocol for non-scientist users of the assay was developed and improved through independent trials with different end-user groups. The assay applicability was verified in a final validation trial with participants without scientific background resulting in 50 percent of the participants successfully detecting S. spallanzanii. Participants rated the ease of use and performance and read-out mostly as easy-to-very easy with overall positive written feedback on its usability for citizen science applications.
The detailed information on the assay development, validation and trials with the non-scientist users has been published in Frontiers in Marine Science journal.
The full study can be accessed here.




Thank you so much for your work. This RPA test for fanworm is amazing, and it’s inspiring to see how you are connecting people with the science. As boaters ourselves, currently underway from Canada to New Zealand, we care deeply about the ocean. But it’s heartbreaking to think that our love for the ocean might harm the very systems that we enjoy so much. Hopefully your efforts will help motivate further development of non-toxic, anti-attachment hull treatments to better protect our boats and our environment.
Dear Brooksie, thank you for your message and for following our updates! It is the best reward and encouragement for us to see support and endorsement from people passionate about the ocean and its health. With the rapid development of new technologies, we see a lot of opportunities to better protect marine systems from human-induced stressors (including incursions of non-native species) and work towards harnessing these exciting technologies within the Marine Biosecurity Toolbox. One of the Programme’s workstreams (Protect) is specifically looking into novel and ecosystem-friendly ways to prevent biofouling accumulation on underwater structures. We’ll be happy to share more updates on these developments soon – please stay tuned!
Anastasija Zaiko,
on behalf of the Marine Biosecurity Toolbox team