Last year over 700 boat owners who have non-trailered boats moored in marinas around Aotearoa…
Experimental insights on effective eDNA/eRNA capture from seawater samples
Although there is still no consensus on a standardized workflow for processing eDNA/eRNA samples, in aquatic systems the common practice is to concentrate waterborne nucleic acids (NAs) via filtration. Yet, the type of filter membranes and pore sizes vary widely from study to study. It is generally assumed that the smallest pore size helps maximizing NA capture from a wide range of genetic material (including sub-cellular particles). However, using finer pore membranes increases the likelihood of clogging and prohibits processing of larger water volumes, thus potentially reducing the chances of detecting rare biodiversity. Alternatively, splitting samples into multiple sub–samples, which invariably increases consumable costs for filtration and downstream analyses, can potentially compromise the integrity of NAs due to increased handling times. This is especially relevant for eRNA which is more prone to degradation. On the other hand, processing large sample volumes with larger pore membranes may not effectively capture smaller particles, and can increase the concentration of inhibitory substances that may suppress molecular signals and decrease target detectability.
Although it has been previously shown that larger pore filters can be as effective as finer pore filters for species detection from aquatic eDNA, more data are needed on the selectivity of different membrane pore sizes towards specific NA types and fractions. Rarely (if at all) do such comparative studies involve technical efficiency assessment to address time/cost inputs in the context of molecular signal recovery. This restricts informed decision-making around an optimized sampling approach (i.e. combination of time effort and maximized signal detection) for addressing a particular research or surveillance question, for example, the detection and monitoring of nuisance organisms, endangered and indicator taxa, and species of ecological, economical and/or cultural importance.
The DETECT and the ECONOMICS & DECISION-SUPPORT teams have conducted an experimental study using a cultured microalgal species as a model for a controlled filter performance comparison combined with a formal efficiency modelling component which, to our knowledge, has not yet been applied in eDNA and eRNA research (Fig. 1).
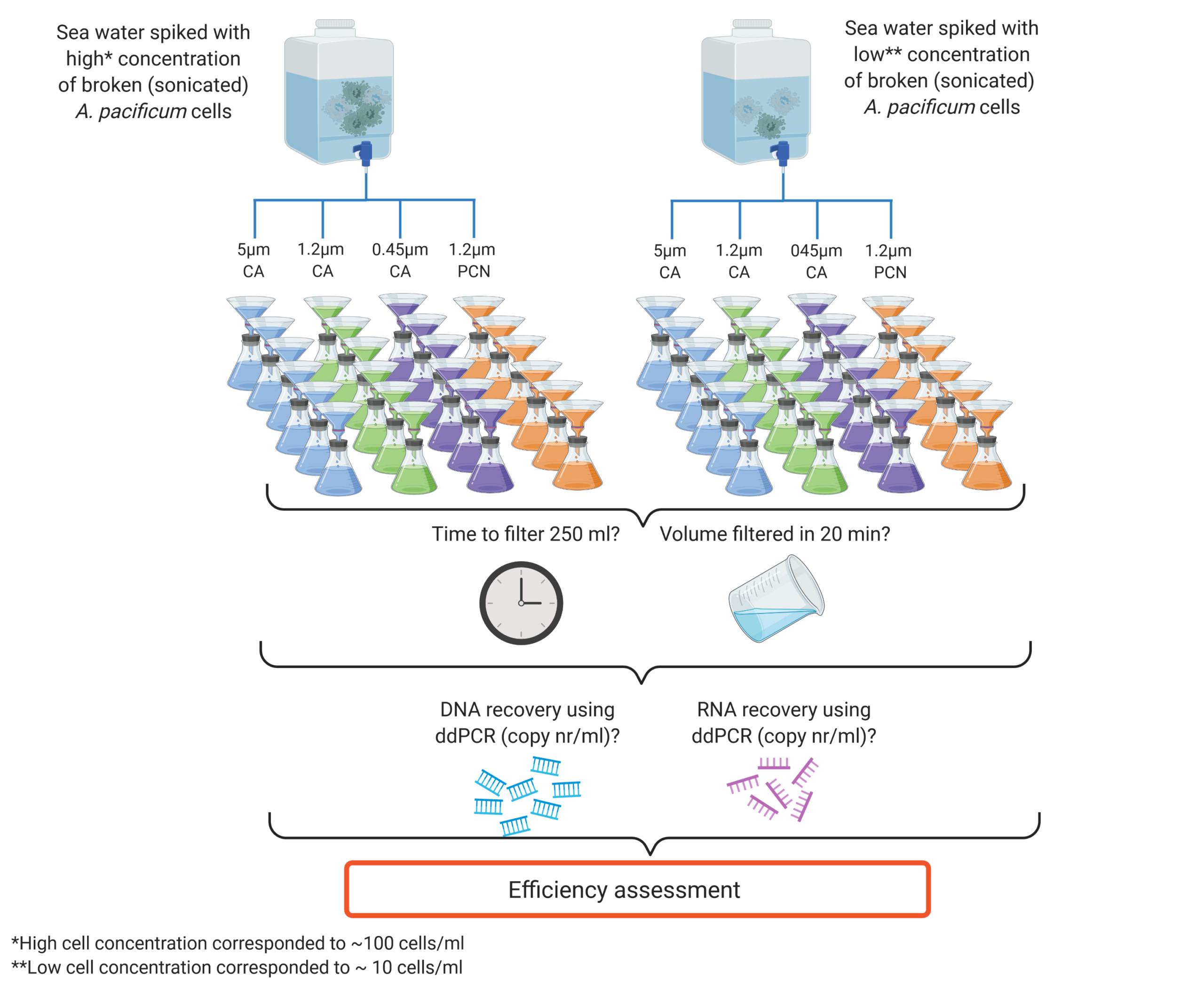
Figure 1: Schematic of the experimental design applied to assess the efficiency of different membranes for eDNA and eRNA capture.
The results showed no statistically significant difference between membrane types for capturing target eDNA signal from intact and partially lysed cell treat–ments. In terms of time effort and volume processed, higher efficiency ratings were obtained with the larger pore size (5 μm) cellulose membranes (Fig. 2). Positively charged nylon demonstrated enhanced capture of naked NAs, and especially eRNA signal, across treatments.
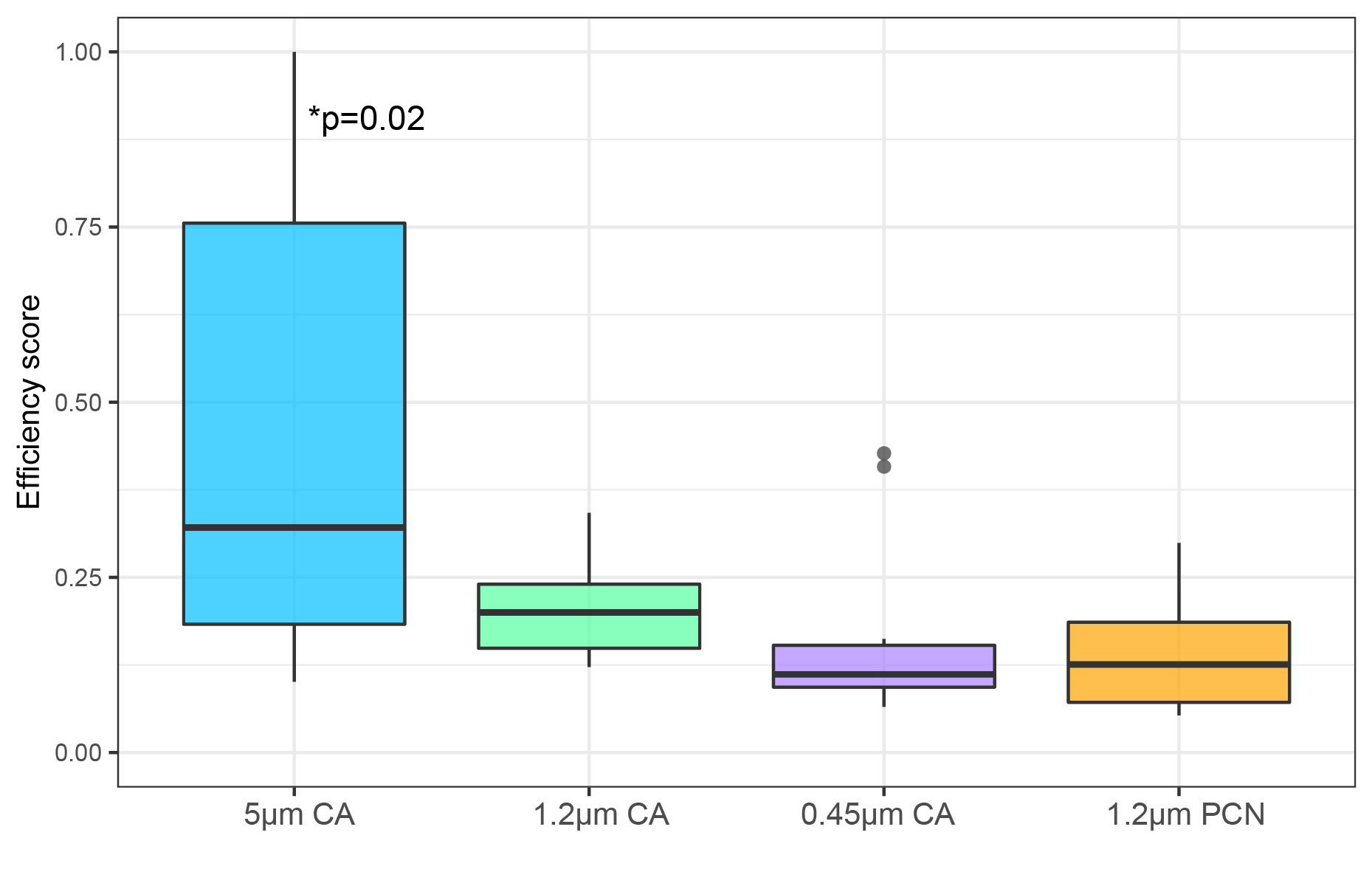
Figure 2: Overall efficiency scores derived from the Data Envelopment Analysis for four tested filter membrane types.
Our findings, recently published in the Methods in Ecology and Evolution journal, support using coarse pore size filters for adequate capture of target NA signal (from both eDNA and eRNA) with less processing time. The evidence that coarse pore-size membranes might perform as well as smaller pore- size filters for capturing different fractions of NAs from water has important practical implications for monitoring and surveil-lance programmes. Efficient sampling of NAs from varied aquatic ecosystems routinely involves concentration of heterogeneously distributed genetic material across large areas or water volumes. This crucial aspect of the workflow is quite challenging, entailing tedious, time and resource-intensive human effort. By using larger pore- size membranes for sample filtration, the recurring problem of rapid clogging can be mitigated, allowing for larger volumes of water to be processed and hence— better recovery of waterborne NAs.




Comments (0)